

Okazaki-type DNA-RNA hybrids are substrates for a number of enzymes.
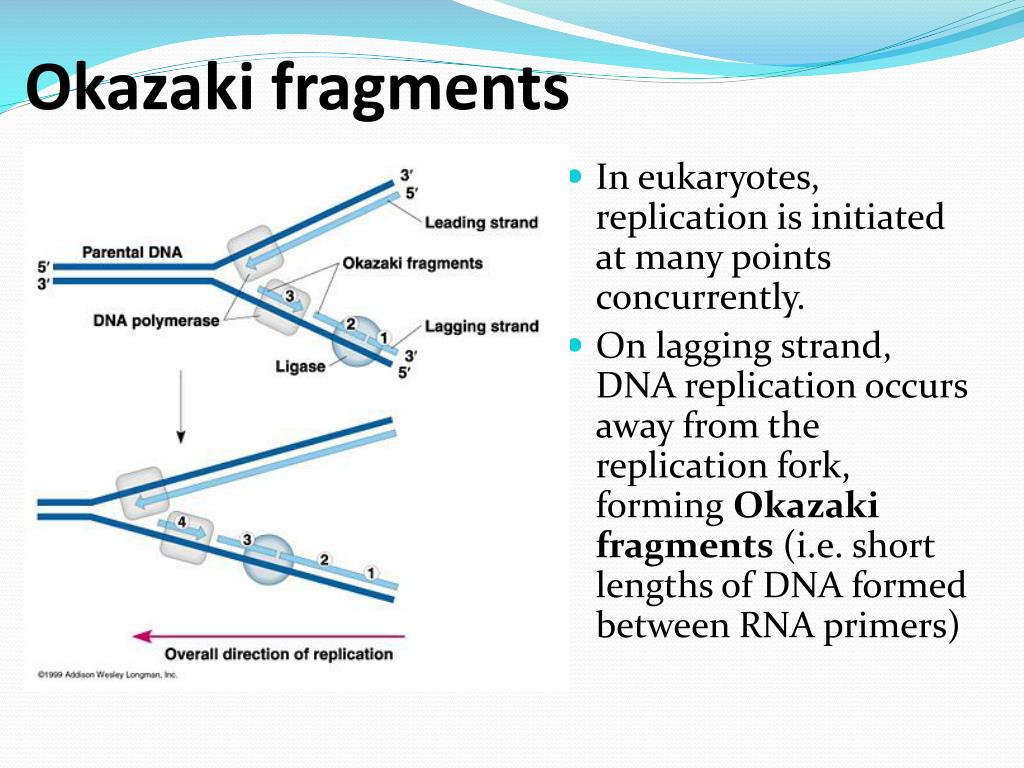
10 residues long) and is then elongated by the DNA polymerase [reviewed in Ogawa and Okazaki (1980)l. This lagging strand is primed by short RNA oligonucleotides laid down by a primase (ca. a 3’-hydroxyl terminus of a primer and cannot initiate new chains de novo (Kornberg, 1974 Okazaki et al., 1967,1968). Ribonucleotides are in italic type in the text. A dash (-) indicates covalently linked residues within the same strand. Symbols: A dot (-) indicates a hydrogen-bonded interaction between bases or strands. * New address: Ribozyme Pharmaceuticals Inc., 2950 Wilderness Place, Boulder, CO 80301. 5 New address: OrganicChemistryLaboratory,Swiss Federal Institute of Technology, CH-8092 Ziirich, Switzerland. * Author to whom correspondence should be addressed. t Atomic coordinates and isotropictemperature factors for the crystal structures have been deposited in the Brookhaven Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (entry numbers 1D87 (G-chimera), 1D88 (A-chimera),and lOFX (Okazaki fragment). was funded by an NIH Fogarty International Research Fellowship. In replication, one DNA strand is copied continuously (the leading strand), while the complementary template strand is replicated in a discontinuous way since both prokaryotic and eukaryotic DNA polymerases require This research was supported by grants from the National Institutes of Health, the AmericanCancerSociety, the National Science Foundation, the Office of Naval Research, and the National Aeronautics and Space Administration.
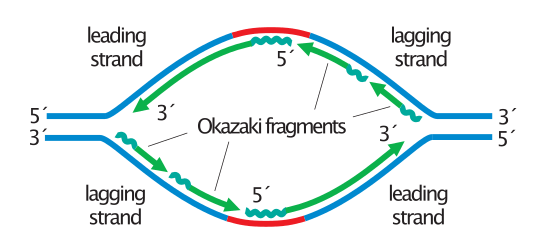
Called an Okazaki fragment, it may be viewed as a short DNA-RNA hybrid duplex covalently linked to a DNA-DNAduplex. Such DNA-RNA chimeric duplexes are formed in replication during DNA synthesis of the lagging strand, where a chimeric DNA-RNA strand is paired with an all- DNA strand. In the other hybrid type, the duplex contains DNAaRNA’ hybrid base pairs in one portion of the molecule and normal DNA-DNA base pairs in the rest of the molecule. This type of hybrid is also present during reverse transcription of viral RNA sequences into DNA. These duplexes were first studied with synthetic polynucleotides (Rich, 1960). In one, formed during transcription of DNA into RNA, the hybrid consists of a homo-DNA and a homo-RNA strand. Two types of hybrid duplexes can be differentiated. Hybrids between DNA and RNA play a crucial role in biological information transfer. In DNA-RNA chimeric duplexes, it is therefore possible that a single RNA residue can drive the conformational equilibrium toward the A-conformation. Moreover, the oligonucleotide does not match any of the sequence characteristics of DNAs usually adopting the A-conformation in the crystalline state (e.g., octamers with short alternating stretches of purines and pyrimidines). Crystals of an all-DNA strand with the same sequence as the self-complementary chimeras show a morphology which is different from those of the chimera crystals. In this molecule the three RNA residues may therefore lock the complete decamer in the A-conformation. CD spectra suggest that the Okazaki fragment assumes an A-conformation in solution as well. The crystalline chimeric duplexes do not show a transition between the DNA A- and B-conformations. Hydrogen bonds between 2’-hydroxyls and 5‘-oxygens or phosphate oxygens, in addition to the previously observed hydrogen bonds to 1’-oxygens of adjacent riboses and deoxyriboses, are observed in the DNA-RNA chimeric duplexes. A number of intramolecular interactions of the ribose 2’-hydroxyl groups contribute to the stabilization of the A-conformation. The crystal structures are non-isomorphous, and the crystallographic environments for the three chimeras are different. The two self-complementary duplexes 2 and 2, as well as the Okazaki fragment d(GGGTATACGC).t(GCG)d(TATACCC), were found to adopt A-type conformations. Biochemistry 1993,32, 3221-3237 322 1 Conformational Influence of the Ribose 2’-Hydroxyl Group: Crystal Structures of DNA-RNA Chimeric Duplexest3t Martin Egli,’,s Nassim Usman,I and Alexander Rich Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139 Received SeptemRevised Manuscript Received DecemABSTRACT: We have crystallized three double-helical DNA-RNA chimeric duplexes and determined their structures by X-ray crystallography at resolutions between 2 and 2.25 A.


 0 kommentar(er)
0 kommentar(er)
